Viral infections are associated with changes in the upper respiratory tract/nasopharyngeal (NP) microbiome. In addition, many studies hypothesize the possibilities of ‘super infections’ due to suppressed immunity during a viral onslaught.
Virulent pathogens can be promoted by microbiota, but commensal microbiota can also suppress the spread of viruses. It has been shown that these interactions affect clinical outcomes.
How does the ongoing COVID-19 (coronavirus disease 2019) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), affect the bacterial microbiome?
In a recent study led by Dr. Amy K Feehan, researchers hypothesized that the COVID-19 status would be associated with a shift in the NP microbiome, especially in hospitalized patients. The results from this study support the dynamic and significant changes to the microbiome in the NP space, especially when patients use a breathing assist device.
Breathing treatments are provided to hospitalized COVID-19 patients. In this study, the researchers examined the community composition in relation to three attributes – COVID-19 status, breathing assistance, and antibiotic use. This study is published in the journal, Applied Microbiology.
“Cross-sectional differences in the microbiome were apparent with SARS-CoV-2 infection, but longitudinal studies are needed to understand the dynamics of viral and breathing treatment modulation of microbes.”
The study
The researchers used shallow shotgun sequencing of a cross-sectional set of medical waste samples from Louisiana, USA, that were confirmed by PCR positive for SARS-CoV-2 across a range of severity (asymptomatic, hospitalization, and death). This study included 79 SARS-CoV-2 positive samples and 20 negative samples (control).
The researchers presented the demographics associated with the samples. For example, information such as inpatient, use of a breathing assist device, COVID-19 severity, smoking status, age, gender, and race.
Using metagenomic sequencing and classification, they assessed the microbiome composition of nasopharyngeal swabs of these SARS-CoV-2 positive and negative individuals.
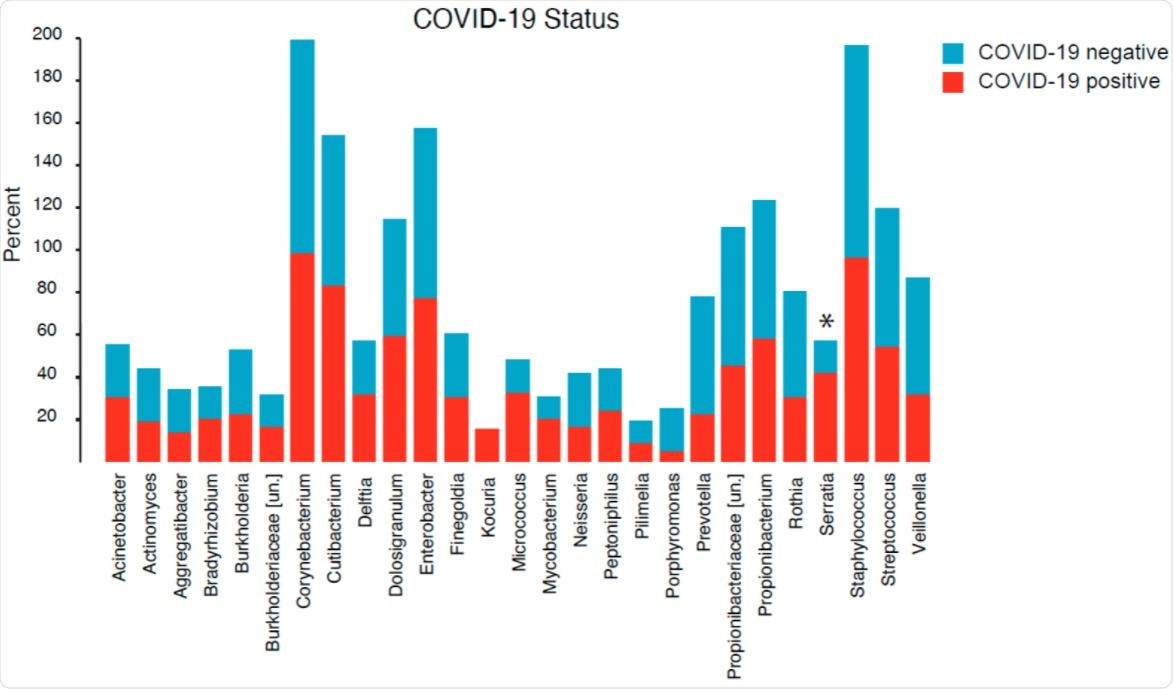
Community composition and differential abundance
They found a total of 202 unique genera across all samples, of which, they identified a total of 27 unique genera. In addition, COVID-19 positivity was associated with increased representation of Serratia, a ubiquitous microorganism. Notably, Serratia marcescens is a recognized causative agent of human diseases, including pneumonia.
The researchers found Serratia, Streptococcus, Enterobacter, Veillonella, Prevotella, and Rothia frequently in COVID-19 patients who used breathing assistive devices.
Also, they reported that two genera: Finegoldia, an opportunistic human pathogen, and Peptoniphilus, typically associated with the gut and vaginal microbiota, were entirely absent in the COVID-19 patients requiring breathing assistance; while present in patients who did not require the breathing assistance.
They noted that the individual relative abundance of Veillonella was similar between groups, and the relative abundance of Finegoldia was clearly higher in the antibiotic usage group.
Species Diversity
They calculated the alpha diversity (diversity within a particular area or ecosystem; species richness) and the beta diversity (difference in diversity of species between ecosystems; species turnover).
Unlike the COVID-19 status, the researchers found that the alpha diversity was different for age status and for individuals who smoked. It is well known that age and smoking affect the microbiome of the gut, skin, and upper respiratory tract.
While they found the beta diversity of the microbiome to be significantly different for those who had COVID-19, used breathing assistive devices, had an inpatient hospital stay, and, also, those who died. Surprisingly, the researchers observed racial differences in beta-diversity of the NP space and emphasized the need for a large-sampled study to evaluate this further.
“The interplay between SARS-CoV-2 infection, breathing treatment, hospital stay, and respiratory microbiomes is complex, and a detailed timeline and/or longitudinal sampling of patients is required to identify causal relationships.”
Summary
While this study does not consider the use of specific medications taken by the individuals due to limitations at the time of data collection, it powerfully highlights the relation between COVID-19 infection and the changes in the microbial communities. It is evident that COVID-19 changes the NP microbiome – in diversity, frequency, and abundance. Additionally, the breathing treatment impacts the type and abundance of the microbial taxa comprising the NP microbiome.
Differences in patient populations, geography, and climate, or even quantity or genomic diversity of the SARS-CoV-2 virus altogether play some role in the NP microbiome.
- Feehan, A.K.; Rose, R.; Nolan, D.J.; Spitz, A.M.; Graubics, K.; Colwell, R.R.; Garcia-Diaz, J.; Lamers, S.L. Nasopharyngeal Microbiome Community Composition and Structure Is Associated with Severity of COVID-19 Disease and Breathing Treatment. Appl. Microbiol. 2021, 1, 177-188. https://doi.org/10.3390/applmicrobiol1020014, https://www.mdpi.com/2673-8007/1/2/14/htm
Posted in: Men's Health News | Medical Research News | Women's Health News | Disease/Infection News
Tags: Antibiotic, Breathing, Coronavirus, Frequency, Genomic, Hospital, immunity, Microbiology, Microbiome, Microorganism, Pandemic, Pathogen, Pneumonia, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Shotgun Sequencing, Skin, Smoking, Syndrome, Vaginal, Virus

Written by
Dr. Ramya Dwivedi
Ramya has a Ph.D. in Biotechnology from the National Chemical Laboratories (CSIR-NCL), in Pune. Her work consisted of functionalizing nanoparticles with different molecules of biological interest, studying the reaction system and establishing useful applications.
Source: Read Full Article



