Hunting for disease clues in the dark matter of our DNA, scientists have reconstructed the chromosomes of patients with an extremely high number of aberrations in their genome that could alter the expression of nearby genes and potentially cause disease. Their results were published in Nature Communications.
“We are only beginning to understand how mutations in the non-coding part might cause disease,” says geneticist Stefan Mundlos, Head of the research group Development & Disease at the Max Planck Institute for Molecular Genetics (MPIMG) and Director of the Institute of Medical and Human Genetics at Charité—Universitätsmedizin Berlin. “This study helps us understand how critical non-coding DNA can be and how genomic rearrangements can bring about diseases.”
Over 20 years ago, Mundlos treated a patient who had a mild intellectual disability. Back then, the techniques used by scientists to peer into a person’s genome were rudimentary. But even using old fashioned karyotyping, Mundlos could see that she had a highly unusual number of aberrations in some of her chromosomes. And yet, she had survived.
The unusual case remained in the back of Mundlos’ mind, and the woman’s cells were frozen in liquid nitrogen until 2019. That’s when Mundlos and post-doctoral researchers Robert Schöpflin and Uirá Souto Melo at MPIMG decided to take a closer look at her chromosomes using newly developed genomic tools.
Knowledge about genetics and its link with disease had expanded by then. Just a decade ago, scientists thought that only mutations in genes that code for proteins cause disease. Such coding genes account for only two percent of the human genome. The remaining 98 percent was termed “junk DNA.”
But with advances in sequencing technology, scientists have learned that even non-coding DNA can be vitally important. “Parts of it can help regulate gene expression and mutations in non-coding regions can lead to disease”, says Melo. Given how little is known about non-coding DNA, some people call it the “dark matter” of our DNA.
“This ‘dark matter’ may be dysfunctional in many diseases,” Melo says. Only 40 percent of patients with genetic diseases ever get a diagnosis because geneticists cannot spot any obvious errors in the coding regions. “We believe that a high fraction of these mutations are in the non-coding regions,” Melo says. “It has to be, otherwise we are missing something very basic in biology.”
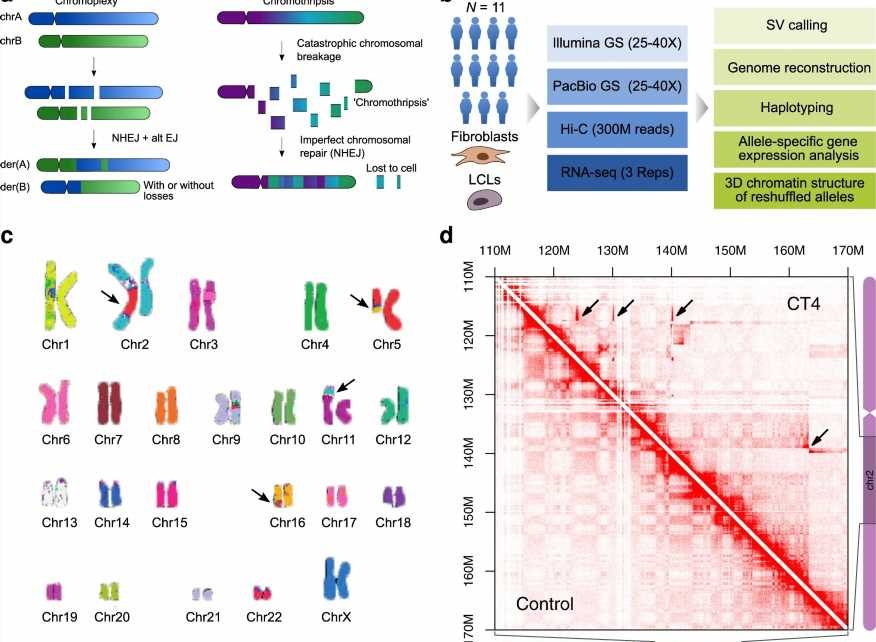
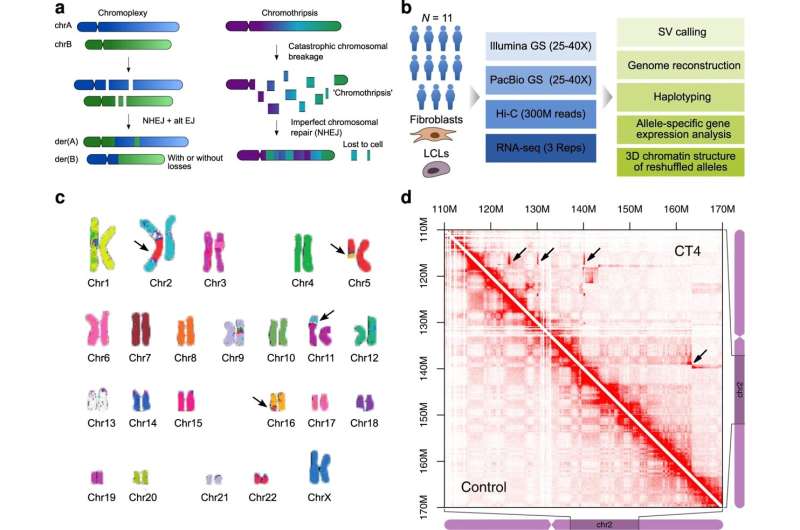
In 2011, scientists reported that sometimes DNA can shatter into pieces in a catastrophic event called “chromothripsis.” The cell’s repair technicians try to reassemble the pieces, but with so many fragments, it’s not surprising that the end product doesn’t always resemble the original. If too many pieces are missing, the cell will die. But if the botched job happens in a “non-coding” region, then the person could potentially survive. Sometimes, patients are fine even with the aberrant DNA. Other times, they exhibit a genetic disease.
But people having such extreme chromosomal rearrangements in all of their cells are rare. Melo says there are only a few reported cases worldwide who have undergone chromothripsis and survived. Mundlos’ patient twenty years ago was one of them. Her DNA became a valuable resource for the team, who were wanting to explore the mysteries of non-coding DNA.
The team mulled over how to analyze her genome. Genome sequencing techniques are advancing rapidly together with knowledge about how DNA, which measures almost two meters stretched out end-to-end, gets compressed into the 0.05-mm-diameter cell nucleus.
During the precise packaging, regions of DNA fold into 3D structures called “topologically associating domains” or TADs. These are complexes of genes and regulatory elements arranged into distinct shapes as determined by their nucleotide sequence.
Melo and his colleagues used a tool called Hi-C combined with genome sequencing to read the 3D structure of these TADs. They could then reconstruct the structure of the patient’s DNA at high resolution. “We were very surprised to detect many previously unknown rearrangements with this method,” Melo says. “The affected individuals had, on average, 28 different genomic rearrangements in the genome.”
The scientists contacted other geneticists who might have known patients with chromothripsis. The team eventually received samples from eleven patients. Ten of them had intellectual disability, while one had no noticeable disorder. Some of them had died, but they had agreed their cells could be used for research purposes.
The scientists found that most of the chromosome aberrations were located in inactive parts of the genome. That’s hypothetically why the people had survived, Melo says. If the mutations had been located in an active region, they would have likely been lethal, or the individuals would have exhibited even more aggressive disorders.
But if the aberrations were in inactive regions, why did many of the patients have an intellectual disability? The scientists found that the shape of the TADs had changed. And this had the potential to affect the expression of genes nearby. In fact, the closer a gene was to the chromosome breakpoint, the more likely it was to be affected.
“This study shows the potential of new sequencing technologies in combination with advanced computer algorithms to understand rare genetic diseases and the organization of our genetic material in the nucleus,” says Martin Vingron, Director at the MPIMG, who was heading the computational efforts for the study.
There are so many chromosomal breaks in chromothripsis that it is difficult to say which of them led to the observed disorder, Vingron says. But as techniques improve, there will soon be more light shed on the dark matter of our genes. “The techniques capable of sequencing every part of the DNA came out only in the last two to three years. We are really at the frontier right now.”
More information:
Robert Schöpflin et al, Integration of Hi-C with short and long-read genome sequencing reveals the structure of germline rearranged genomes, Nature Communications (2022). DOI: 10.1038/s41467-022-34053-7
Journal information:
Nature Communications
Source: Read Full Article