A recent study shows how a novel and integrated lab-on-a-chip platform can represent a rapid, affordable, and exact molecular diagnostic tool for detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The paper is currently available for free on the medRxiv* preprint server while it undergoes peer review.
As a response to the ongoing coronavirus disease (COVID-19) pandemic and evident disparities of vaccination coverage in low- and middle-income countries, it is pivotal to adhere to a widespread testing and screening program for surveillance and control of infections in regions where there are limited medical resources.
The gold standard for diagnosing SARS-CoV-2 infection is the use of the real-time reverse transcription-polymerase chain reaction (RT-qPCR) to detect viral ribonucleic acid (RNA) in specimens from the respiratory tract. But although the method is highly sensitive and specific, it necessitates expensive equipment and highly skilled personnel.
Several point-of-care RNA detection technologies circumvent the need for expensive instruments and simultaneously use reverse transcription and isothermal amplification; one notable example is loop-mediated isothermal amplification (LAMP) which is slowly gaining prominence for many different infectious diseases.
And then there are CRISPR-Cas-assisted SARS-CoV-2 detection assays, which are viewed as transformative methods for point-of-care COVID-19 diagnostics. However, they currently lack streamlined sample preparation and integration within the automated, portable system.
This new manuscript, first-authored by Dr. Bongkot Ngamso from the University of Hull in the United Kingdom, demonstrated the manual operation of a microfluidic-based CRISPR device as a molecular COVID-19 diagnostic tool by a semi-trained operator in resource-limited laboratories in sub-Saharan Africa (such as Kenya).
A combination of microfluidics and CRISPR-Cas
The researchers combined a microfluidic technique known as immiscible filtration assisted by surface tension (IFAST) with the recent developments in CRISPR-Cas12-based sensing, resulting in a sensitive, cost-effective, target-specific, and completely integrated device for COVID-19 diagnostics.
The device was dubbed ‘IFAST-CRISPR’. It streamlined sample preparation to enable rapid isolation and concentration of RNA directly from nasopharyngeal swab samples or saliva specimens, followed by CRISPR-Cas-assisted detection with lateral flow readout.
In a nutshell, the use of adequate functionalized magnetic particles permits isolation and purification of a magnetically responsive analyte directly from complex matrices, which is a technology that could be used in the future for other infectious diseases.

IFAST-CRISPR device for SARS-CoV-2 detection. (A) Design and (B) photograph of the IFAST-CRISPR device. Chamber 1 = sample + GuHCl + silica paramagnetic beads; chambers 2, 4, 6, 8 = mineral oil; chamber 7 = RT-LAMP reagent; chamber 9 = CRISPR-Cas12 reagent. (C) IFAST-CRISPR device detects SARS-CoV-2 viral RNA from unprocessed nasopharyngeal (NP) swab or saliva sample in a 1 h sample-to-answer workflow. Step 1: RNA is extracted from a sample via silica paramagnetic beads and 5 M GuHCl. Step 2: The MB-isolated RNA is in vitro transcribed and amplified into DNA amplicons via RT-LAMP. Step 3: The hybridization of the targeted DNA sequence activates the gRNA-Cas12a complex to digest ssDNA probe, thereby producing a test line (T) on the lateral flow strip which can be visualized by the naked eye. (D) Principle of the lateral flow readouts for SARS-CoV-2 detection. Control line (C) appears from the intact FAM-biotinylated ssDNA reporter. Test line (T) is present from cleaved ssDNA reporter following target dsDNA-gRNA hybridization.
High sensitivity and specificity
By combining the aforementioned LAMP with CRISPR-Cas12 assays targeting the nucleoprotein gene of SARS-CoV-2, the researchers achieved visual identification of more than 470 viral copies per mL in 45 minutes – without any cross-reactivity towards seasonal coronaviruses or influenza.
Furthermore, on-chip assays revealed the ability to detect and isolate SARS-CoV-2 from one thousand genome copies of replication-deficient viral particles in one hour, with the potential to be additionally optimized and refined.
In short, this affordable, simple, and yet highly integrated platform showed sensitivity and specificity characteristics comparable to the much more expensive gold standard that is RT-qPCR – requiring only a simple heating source.
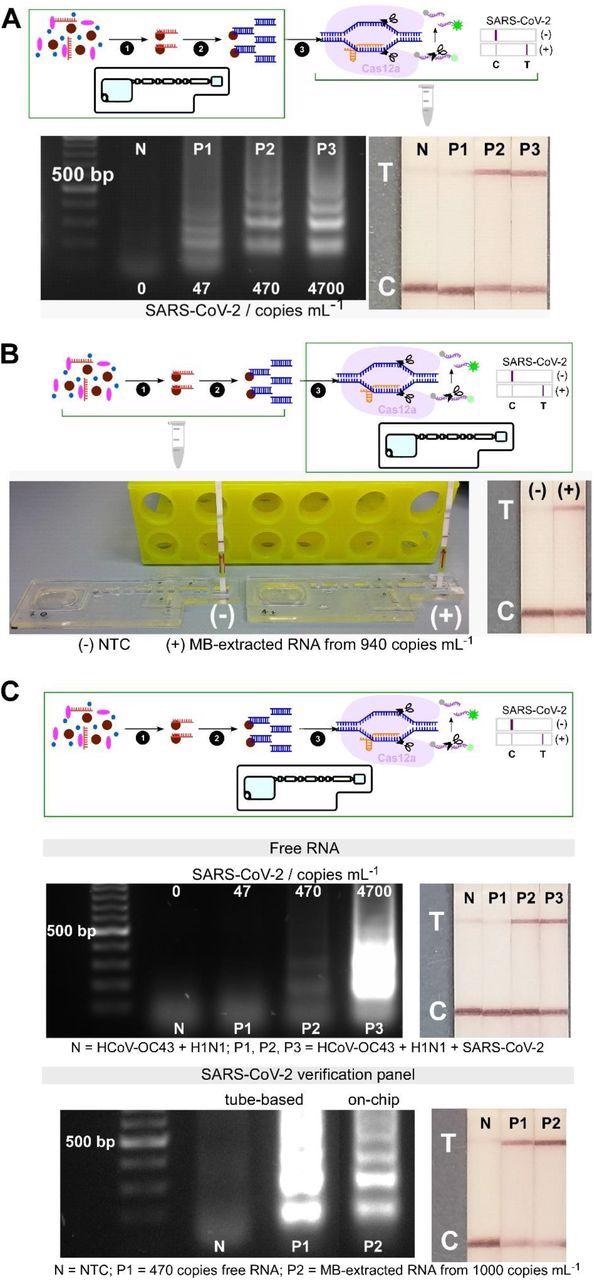
Analytical validation of individual and combined on-chip assays. (A) On-chip RNA extraction, followed by RT-LAMP assays; gel electrophoresis results showing target dsDNA being amplified from MB-extracted RNA from ≥ 470 copies mL-1 initial concentrations, confirmed by test lines on lateral flow test strips (n=2). (B) On-chip CRISPR-Cas12 assays of amplicons from tube-based RNA extraction and RT-LAMP – collateral cleavage of lateral flow ssDNA reporters following the hybridization between the gRNA and dsDNA target showing a test line in positive sample from MB-extracted RNA (n=1). (C) On-chip integrated steps of RNA extraction, RT-LAMP and CRISPR-Cas-assisted detection from samples containing free genomic SARS-CoV-2 RNA (in a mixture containing HCoV-OC43 and H1N1 RNAs, n=2), and from viral particles containing SARS-CoV-2 genome (SARS-CoV-2 verification panel, n=1).
A future of diagnostics
The adaptability of this platform can definitely be implemented for CRISPR-Cas-based detections of other pathogens, which is a great promise as a future addition to the point-of-care diagnostic armamentarium, particularly in resource-limited and decentralized regions of low- and middle-income countries.
“The platform required only a basic heating source such as simple incubators or hot plates which are usually available in most laboratories in low-resource settings, without the need for costly or specialized instruments”, emphasize study authors in this medRxiv paper.
Further research on multiplexing and direct interfacing of the easily obtainable Swan-brand cigarette filter for saliva specimen collection could provide a steadfast workflow for COVID-19 diagnostics from saliva samples amenable for low-resource settings.
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Ngamsom, B. et al. (2021). An integrated lab-on-a-chip device for RNA extraction, amplification and CRISPR-Cas12a-assisted detection for COVID-19 screening in resource-limited settings. medRxiv. https://doi.org/10.1101/2022.01.06.22268835, https://www.medrxiv.org/content/10.1101/2022.01.06.22268835v1
Posted in: Device / Technology News | Medical Science News | Medical Research News | Disease/Infection News
Tags: Analyte, CHIP, Cigarette, Coronavirus, Coronavirus Disease COVID-19, covid-19, CRISPR, Diagnostic, Diagnostics, DNA, Electrophoresis, Eye, Gel Electrophoresis, Gene, Genome, Genomic, H1N1, Hybridization, in vitro, Incubators, Infectious Diseases, Influenza, Lab-on-a-Chip, Microfluidics, Nasopharyngeal, Pandemic, Polymerase, Polymerase Chain Reaction, Research, Respiratory, Ribonucleic Acid, RNA, RNA Extraction, Sample Preparation, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Transcription

Written by
Dr. Tomislav Meštrović
Dr. Tomislav Meštrović is a medical doctor (MD) with a Ph.D. in biomedical and health sciences, specialist in the field of clinical microbiology, and an Assistant Professor at Croatia's youngest university – University North. In addition to his interest in clinical, research and lecturing activities, his immense passion for medical writing and scientific communication goes back to his student days. He enjoys contributing back to the community. In his spare time, Tomislav is a movie buff and an avid traveler.
Source: Read Full Article



